-
Notifications
You must be signed in to change notification settings - Fork 51
/
block10_latticeNittyGritty.rmd
284 lines (218 loc) · 19.4 KB
/
block10_latticeNittyGritty.rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
Backfilling some technical info on `lattice`
========================================================
```{r include = FALSE}
## I format my code intentionally!
## do not re-format it for me!
opts_chunk$set(tidy = FALSE)
## sometimes necessary until I can figure out why loaded packages are leaking
## from one file to another, e.g. from block91_latticeGraphics.rmd to this file
if(length(yo <- grep("gplots", search())) > 0) detach(pos = yo)
if(length(yo <- grep("gdata", search())) > 0) detach(pos = yo)
if(length(yo <- grep("gtools", search())) > 0) detach(pos = yo)
```
Now that you have made many `lattice` plots, let's fill in some technical details that will help you become even more effective in your use of `lattice`.
### Optional getting started advice
*Ignore if you don't need this bit of support.*
This is one in a series of tutorials in which we explore basic data import, exploration and much more using data from the [Gapminder project](http://www.gapminder.org). Now is the time to make sure you are working in the appropriate directory on your computer, perhaps through the use of an [RStudio project](block01_basicsWorkspaceWorkingDirProject.html). To ensure a clean slate, you may wish to clean out your workspace and restart R (both available from the RStudio Session menu, among other methods). Confirm that the new R process has the desired working directory, for example, with the `getwd()` command or by glancing at the top of RStudio's Console pane.
Open a new R script (in RStudio, File > New > R Script). Develop and run your code from there (recommended) or periodicially copy "good" commands from the history. In due course, save this script with a name ending in .r or .R, containing no spaces or other funny stuff, and evoking "technical background" and "lattice".
### Load the Gapminder data, drop Oceania, load packages
Assuming the data can be found in the current working directory, this works:
```{r, eval=FALSE}
gDat <- read.delim("gapminderDataFiveYear.txt")
```
Plan B (I use here, because of where the source of this tutorial lives):
```{r}
## data import from URL
gdURL <- "http://www.stat.ubc.ca/~jenny/notOcto/STAT545A/examples/gapminder/data/gapminderDataFiveYear.txt"
gDat <- read.delim(file = gdURL)
```
Basic sanity check that the import has gone well:
```{r}
str(gDat)
```
Drop Oceania, which only has two continents
```{r}
## drop Oceania
jDat <- droplevels(subset(gDat, continent != "Oceania"))
str(jDat)
```
Load the `lattice` and `plyr` packages.
```{r}
library(lattice)
library(plyr)
```
### High level `lattice` functions actually return objects
```{r}
myAwesomePlot <-
xyplot(lifeExp ~ gdpPercap | continent, jDat,
scales = list(x = list(log = 10, equispaced.log = FALSE)),
type = c("p", "smooth"), grid = TRUE, col.line = "darkorange", lwd = 4)
```
Hello? Where is the plot? Our previous plots have been appearing in a graphics device on your screen (and inserted into this document) because, by default, the `print()` method for objects of class `trellis` has been dispatched on the return value of our high-level `lattice` commands. If you make an assignment, as we did above, or if the command is wrapped inside a function, inside a `plyr` call, or inside a loop, automatic printing will not occur. To explicitly print the figure use `print()` or `plot()`.
```{r}
print(myAwesomePlot)
```
It is possible to do other things to a `trellis` object, such as `update()` or `summary()` but I rarely do.
### Putting more than one `lattice` plot on a "page"
Another reason to assign the object and call `print()` explicitly is to exploit additional arguments (read the help on `print.trellis()` to see them all). The ones you will use most facilitate the placement of more than 1 figure on a page; the arguments are`pos =` and `split =`. Here we use `print(..., pos =...)` to arrange two plots together.
```{r fig.width = 12}
myOtherPlot <- stripplot(lifeExp ~ reorder(continent, lifeExp),
subset(jDat, subset = year %in% c(1952, 1977, 2007)),
groups = year, auto.key = list(reverse.rows = TRUE),
jitter.data = TRUE, type = c("p", "a"), fun = median)
print(myAwesomePlot, pos = c(0, 0, 0.52, 1), more = TRUE)
print(myOtherPlot, pos = c(0.48, 0, 1, 1))
```
The `split =` argument is also useful for this purpose but I prefer `pos =` because I can specify a wee bit of overlap to reduce whitespace between the plots. In the calls above, note that the lefthand plot runs from the far left to just past the halfway mark and the right hand plot begins just before halfway and ends at the far right.
### Extended formula interace of `lattice`
You are probably beginning to realize that data in a tall or long form is easier to plot than data in a short or wide form. Sadly, exactly the opposite is true for creating human-facing tables. So a skilled data analyst is adept at reshaping. That topic is covered more here (link to future reshaping topic). Feeling lazy? `lattice` does offer one trick that may let you get away with short/wide data: the extended formula interface.
Consider one of our [data aggregation tasks](hw03_dataAggregation.html): looking at different measures of spread for life expectancy over time, across different continents.
```{r}
lifeExpSpread <- ddply(jDat, ~ continent + year, summarize,
sdGdpPercap = sd(gdpPercap), iqrGdpPercap = IQR(gdpPercap),
madGdpPercap = mad(gdpPercap))
head(lifeExpSpread)
```
In our aggregated result `lifeExpSpread`, each measure of spread exists as its own variable. Let's say we want to plot those over time, distinguishing the different measures via color and superposition or by multi-panel conditioning, possibly depicting continent as well. To use the native `lattice` functionality for superposition or conditioning, we would need to reshape the data. The variables `sdGdpPercap`, `iqrGdpPercap`, and `madGdpPercap` would need to be catenated and a parallel factor created to indicate `sd` vs `iqr` vs `mad`. If you have no other need for such reshaped data, you can bypass its creation by using the extended formula interface.
```{r fig.show='hold', out.width='50%'}
## cheap trick? using lattice's extended formula interface to avoiding reshaping
xyplot(sdGdpPercap + iqrGdpPercap + madGdpPercap ~ year, lifeExpSpread,
subset = continent == "Africa", type = "b", ylab = "measure of spread",
auto.key = list(x = 0.07, y = 0.85, corner = c(0, 1)))
xyplot(sdGdpPercap + iqrGdpPercap + madGdpPercap ~ year, lifeExpSpread,
group = reorder(continent, sdGdpPercap), layout = c(3, 1),
type = "b", ylab = "measure of spread",
auto.key = list(x = 0.35, y = 0.85, corner = c(0, 1),
reverse.rows = TRUE))
```
In the first figure, we focus on one continent. But we achieve an effect similar to the use of the `group =` argument by specifying multiple terms as primary variables, separated by plus `+` signs. The reshaping described above is carried out temporarily by `lattice`; in particular, a virtual factor is created to distinguish `sd` vs `iqr` vs `mad`. In the second figure, we explicitly request `group = continent` and so the different measures of spread are parcelled out to different panels, as if we had properly requested multi-panel conditioning on the virtual factor.
If you want the virtual factor to be treated as a conditioning factor, the explicit way to request is to set `outer = TRUE`. Above this is achieved as a side effect of specifying `continent` as the grouping variable. Here's a more useful, typical example of how this works:
```{r}
xyplot(sdGdpPercap + iqrGdpPercap + madGdpPercap ~ year, lifeExpSpread,
subset = continent == "Africa", type = "b", ylab = "measure of spread",
outer = TRUE, layout = c(3, 1), aspect = 1)
```
Read more in section 10.2 "The extended formula interface" in Sarkar's `lattice` book.
### High level `lattice` functions have panel functions at their heart
Functions like `stripplot()` and `xyplot()` are examples of *high level* `lattice` functions. Table 1.1 in Sarkar's `lattice` book lists 15 high level functions.
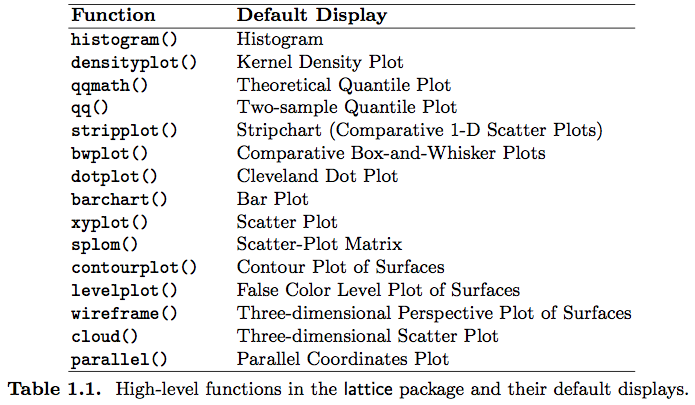
We've pointed out many of the useful features in common to these functions: the `data =` and `subset =` arguments for specifying input data, the formula interface (which supports multipanel conditioning), and the `group =` and `type =` arguments. All high level functions are wrappers around lots of smaller functions that do the grunt work like set up layouts and coordinate systems, draw axes, and add graphical elements. The high level functions can be organized into groups and, within these groups, there is a lot of shared infrastructure with respect to the inner workings. It can simplify your life to understand this, so you can transfer knowledge and tricks from one function to another. Here are the functions you will use most, organized into groups:
1. `densityplot()`, `histogram()`
2. `xyplot()`, `stripplot()`, `bwplot()`, `dotplot()`, `barchart()`
3. `levelplot()`, `contourplot()`
4. `splom()`, `parallelplot()` (was `parallel()`)
Of all the lower level functions wrapped up inside a high level function, the most important one is the __panel function__, which does the actual plotting. Every `lattice` plot consists of at least one panel; in the case of multi-panel conditioning via e.g., `y ~ x | z`, there will be one panel per level of `z`. The input data is chopped into pieces called __packets__, one per panel, very much like the splitting done in `plyr::ddply()`. The panel function then plots the data from one packet into its associated panel.
Each high level function has a default panel function and, by convention, its name is `panel.` followed by the name of the high level function. When simple customizations aren't enough, the first thing to do is to read the help on the default panel function. Seriously, right now go read the help on `panel.xyplot()`! This is where you will gain deeper knowledge of the arguments available for customization. Even though these are arguments of the panel function itself, you can often set them in a high level function call, and they will be passed through due to the use of the special `...` argument (read more about that [here](block06_functions.html). In fact, you've already seen examples like this:
```{r eval = FALSE}
xyplot(lifeExp ~ gdpPercap, jDat,
grid = TRUE,
scales = list(x = list(log = 10, equispaced.log = FALSE)),
type = c("p", "smooth"), col.line = "darkorange", lwd = 3)
```
Call up the help file on `xyplot()` and search it for arguments `col.line` and `lwd`. You will not find them (OK, `lwd` is mentioned in the documentation about keys but that doesn't count). But you *will* find them in the help for `panel.xyplot()`. __Learn this lesson well.__ It's how you can take your `lattice` plots to the next level without too much pain.
### Customizing the panel function
Sometimes the customization you need will require you to modify the panel function. Relax! It's really not that hard and is very rewarding. We will walk through a typical process for writing a custom panel function 'on the fly'.
I present a modified excerpt from my development process for creating a single year "still snapshot" of a typical Gapminder moving bubble chart. I load a souped-up version of the Gapminder data you know and love that [1] includes some stunning country colors and; [2] is sorted by country size so little countries aren't hidden by big ones. Furthermore, we work with a subset of the data for just two years.
```{r}
gdURL <- "http://www.stat.ubc.ca/~jenny/notOcto/STAT545A/examples/gapminder/data/gapminderWithColorsAndSorted.txt"
kDat <- read.delim(file = gdURL, as.is = 7) # protect color
str(kDat)
jYear <- c(1952, 2007)
yDat <- subset(kDat, year %in% jYear)
str(yDat)
```
First start with a high level function call that works, but is missing some of the cool features you want. Crawl before you walk. Walk before you run.
```{r}
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)))
```
My first step is to specify the panel function explicitly but, of course, to specify the default. Do you think pilots switch off auto-pilot in the middle of a delicate maneuver? No, they prefer to take manual control when the plane is flying smoothly at 30,000 feet in clear skies. You will see no change in the figure, which is exactly what we want at this point.
```{r}
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
panel = panel.xyplot)
```
Now I write a custom panel function 'on the fly' but the function body just consists of the defaul panel function. Any arguments normally passed to the panel function are passed straight through to `panel.xyplot()`. I omit the plot because there is no visible change, which is exactly what I want right now.
```{r eval = FALSE}
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
panel = function(...) {
panel.xyplot(...)
})
```
Now I modify the custom panel function to have arguments `x`, `y`, and `...`. If you read the help on `panel.xyplot()`, you will notice that `x` and `y` are the first two formal arguments and they do not have default values. This is why, if I want to specify any of the other arguments, I must explicitly specify `x` and `y`. Below, I take whatever would have normally been given to the panel function as `x` and `y` and am passing them straight through. I omit the plot because there is no visible change, which is exactly what I want right now.
```{r eval = FALSE}
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
panel = function(x, y, ...) {
panel.xyplot(x, y, ...)
})
```
Now I take control of the plotting symbol: I make it an empty circle, which happens to be symbol 21. Conceptually, I scale the radius of the circle so that the area will reflect the country's population. The logic and testing of this mapping of population into circle size was done elsewhere and required some thought and care, FYI. The mechanics are a bit tricky: The term `cex` (think "character expansion) is conventionally used in R as an argument controlling font or symbol size. That's the dial I need to turn to change circle size. I set the `cex =` argument of the high level `xyplot()` call to a vector corresponding to the rows of the input data `yDat`. Since `xyplot()` doesn't have a `cex =` argument, it passes that down to the panel function, which is `panel.xyplot()` in this case, which does have a `cex =` argument. Then, inside the panel function, I consult the `subscripts` argument to learn which rows of the original input data are in the current packet. I can then index `cex` by these subscripts to get the corect radii and draw the circles. Phew.
```{r}
jCexDivisor <- 1500 # arbitrary scaling constant
jPch <- 21
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
cex = sqrt(yDat$pop/pi)/jCexDivisor,
panel = function(x, y, ..., cex, subscripts) {
panel.xyplot(x, y, cex = cex[subscripts], pch = jPch, ...)
})
```
Now I fill the circles with the pre-chosen country colors and explicitly set the circle outlines to a nice tasteful grey. The fill color mechanics mirror those used for setting the circle size.
```{r}
jDarkGray <- 'grey20'
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
cex = sqrt(yDat$pop/pi)/jCexDivisor, fill.color = yDat$color,
col = jDarkGray,
panel = function(x, y, ..., cex, fill.color, subscripts) {
panel.xyplot(x, y, cex = cex[subscripts],
pch = jPch, fill = fill.color[subscripts], ...)
})
```
Since I've come this far, I will go ahead and add a key to link the continents to colors (or color families, here). This has nothing to do with panel functions!
```{r}
gdURL <- "http://www.stat.ubc.ca/~jenny/notOcto/STAT545A/examples/gapminder/data/gapminderContinentColors.txt"
continentColors <- read.delim(file = gdURL, as.is = 3) # protect color
continentKey <-
with(continentColors,
list(x = 0.95, y = 0.05, corner = c(1, 0),
text = list(as.character(continent)),
points = list(pch = jPch, col = jDarkGray, fill = color)))
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
cex = sqrt(yDat$pop/pi)/jCexDivisor, fill.color = yDat$color,
col = jDarkGray, key = continentKey,
panel = function(x, y, ..., cex, fill.color, subscripts) {
panel.xyplot(x, y, cex = cex[subscripts],
pch = jPch, fill = fill.color[subscripts], ...)
})
```
To conclude, we present our starting and ending plots side by side.
```{r fig.show='hold', out.width='50%'}
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10)))
xyplot(lifeExp ~ gdpPercap | factor(year), yDat, aspect = 2/3,
grid = TRUE, scales = list(x = list(log = 10, equispaced.log = FALSE)),
cex = sqrt(yDat$pop/pi)/jCexDivisor, fill.color = yDat$color,
col = jDarkGray, key = continentKey,
panel = function(x, y, ..., cex, fill.color, subscripts) {
panel.xyplot(x, y, cex = cex[subscripts],
pch = jPch, fill = fill.color[subscripts], ...)
})
```
The differences in the code are really not that huge, considering how different the output is.
### Student Q & A
Q: What if you want to plot 2 quantitative variables against another BUT they are on completely different scales? We got kind of lucky above with the different measures of spread -- there was considerable overlap in their observed values and therefore the extended formula interface and a single y-axis "just worked".
A: How to do this is demonstrated in a few places in Sarkar's `lattice` book. This requires these manipulations:
- rescaling at least one variable, to get it on the same scale as the other MANDATORY
- adding an axis for the rescaled variable STRONGLY ENCOURAGED
- distributing `type =` across the multiple primary variables via `distribute.type = TRUE` DO IF NEEDED
See section 5.3.1 (p. 79 - 82) and section 8.4.2 (p. 148 - 150).
### References
Lattice: Multivariate Data Visualization with R [available via SpringerLink](http://ezproxy.library.ubc.ca/login?url=http://link.springer.com.ezproxy.library.ubc.ca/book/10.1007/978-0-387-75969-2/page/1) by Deepayan Sarkar, Springer (2008) | [all code from the book](http://lmdvr.r-forge.r-project.org/) | [GoogleBooks search](http://books.google.com/books?id=gXxKFWkE9h0C&lpg=PR2&dq=lattice%20sarkar%23v%3Donepage&pg=PR2#v=onepage&q=&f=false)
My step-by-step development of a custom panel function was heavily inspired by ["Lattice Tricks for the Power UseR"](http://www.r-project.org/conferences/useR-2007/program/presentations/sarkar.pdf), a talk by Deepayan Sarkar at the 2007 UseR! conference.
<div class="footer">
This work is licensed under the <a href="http://creativecommons.org/licenses/by-nc/3.0/">CC BY-NC 3.0 Creative Commons License</a>.
</div>