-
Notifications
You must be signed in to change notification settings - Fork 51
/
block14_colors.rmd
294 lines (226 loc) · 19.4 KB
/
block14_colors.rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
Using colors in R
========================================================
```{r include = FALSE}
library(knitr)
## I format my code intentionally!
## do not re-format it for me!
opts_chunk$set(tidy = FALSE)
## sometimes necessary until I can figure out why loaded packages are leaking
## from one file to another, e.g. from block91_latticeGraphics.rmd to this file
if(length(yo <- grep("gplots", search())) > 0) detach(pos = yo)
if(length(yo <- grep("gdata", search())) > 0) detach(pos = yo)
if(length(yo <- grep("gtools", search())) > 0) detach(pos = yo)
```
### Optional getting started advice
*Ignore if you don't need this bit of support.*
This is one in a series of tutorials in which we explore basic data import, exploration and much more using data from the [Gapminder project](http://www.gapminder.org). Now is the time to make sure you are working in the appropriate directory on your computer, perhaps through the use of an [RStudio project](block01_basicsWorkspaceWorkingDirProject.html). To ensure a clean slate, you may wish to clean out your workspace and restart R (both available from the RStudio Session menu, among other methods). Confirm that the new R process has the desired working directory, for example, with the `getwd()` command or by glancing at the top of RStudio's Console pane.
Open a new R script (in RStudio, File > New > R Script). Develop and run your code from there (recommended) or periodicially copy "good" commands from the history. In due course, save this script with a name ending in .r or .R, containing no spaces or other funny stuff, and evoking "colors".
### Load the Gapminder data
Assuming the data can be found in the current working directory, this works:
```{r, eval=FALSE}
gDat <- read.delim("gapminderDataFiveYear.txt")
```
Plan B (I use here, because of where the source of this tutorial lives):
```{r}
## data import from URL
gdURL <- "http://www.stat.ubc.ca/~jenny/notOcto/STAT545A/examples/gapminder/data/gapminderDataFiveYear.txt"
gDat <- read.delim(file = gdURL)
```
Basic sanity check that the import has gone well:
```{r}
str(gDat)
```
### Change the default plotting symbol to a solid circle
The color demos below will be more effective if the default plotting symbol is a solid circle. We limit ourselves to base R graphics in this tutorial, therefore we use `par()`, the function that queries and sets base R graphical parameters. In an interactive session or in a plain R script, do this:
```{r eval = FALSE}
## how to change the plot symbol in a simple, non-knitr setting
opar <- par(pch = 19)
```
Technically, you don't need to make the assignment, but it's a good practice. We're killing two birds with one stone:
1. Changing the default plotting symbol to a filled circle, which has code 19 in R. (Below I link to some samplers showing all the plotting symbols, FYI.)
2. Storing the pre-existing and, in this case, default graphical parameters in `opar`.
When you change a graphical parameter via `par()`, the original values are returned and we're capturing them via assignment to `opar`. At the very bottom of this tutorial, we use `opar` to restore the original state.
Big picture, it is best practice to restore the original, default state of hidden things that affect an R session. This is polite if you plan to inflict your code on others. Even if you live on an R desert island, this practice will prevent you from creating maddening little puzzles for yourself to solve in the middle of the night before a deadline.
Because of the way figures are handled by `knitr`, it is more complicated to change the default plotting symbol throughout, e.g., an R Markdown document. To see how I've done it, check out a hidden chunk around here in the [source of this page](https://github.com/jennybc/STAT545A/blob/master/block14_colors.rmd).
```{r include = FALSE}
## see ch. 10 Hooks of Xie's knitr book
knit_hooks$set(setPch = function(before, options, envir) {
if(before) par(pch = 19)
})
opts_chunk$set(setPch = TRUE)
```
### Basic color specification and the default palette
I need a small well-behaved excerpt from the Gapminder data for demonstration purposes. I randomly draw 8 countries, keep their data from 2007, and sort the rows based on GDP per capita. Meet `jDat`.
```{r echo = FALSE}
## take a random sample of countries
nC <- 8
jYear <- 2007
set.seed(1903)
countriesToKeep <- as.character(sample(levels(gDat$country), size = nC))
jDat <-
droplevels(subset(gDat, country %in% countriesToKeep & year == jYear))
jDat <- jDat[order(jDat$gdpPercap), ]
#str(jDat)
```
```{r}
jDat
```
A simple scatterplot, using `plot()` from the base package `graphics`.
```{r}
jXlim <- c(460, 60000)
jYlim <- c(47, 82)
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
main = "Start your engines ...")
```
You can specify color explicitly by name by supplying a character vector with one or more color names (more on those soon). If you need a color for 8 points and you input fewer, recycling will kick in. Here's what happens when you specify one or two colors via the `col =` argument of `plot()`.
```{r fig.show='hold', out.width='50%'}
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = "red", main = 'col = "red"')
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = c("blue", "orange"), main = 'col = c("blue", "orange")')
```
You can specify color explicitly with a small positive integer, which is interpreted as indexing into the current palette, which can be inspected via `palette()`. I've added these integers and the color names as labels to the figures below. The default palette contains 8 colors, which is why we're looking at data from eight countries. The default palette is ugly.
```{r fig.show='hold', out.width='50%'}
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = 1:nC, main = paste0('col = 1:', nC))
with(jDat, text(x = gdpPercap, y = lifeExp, pos = 1))
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = 1:nC, main = 'the default palette()')
with(jDat, text(x = gdpPercap, y = lifeExp, labels = palette(),
pos = rep(c(1, 3, 1), c(5, 1, 2))))
```
You can provide your own vector of colors instead. I am intentionally modelling best practice here too: if you're going to use custom colors, store them as an object in exactly one place, and use that object in plot calls, legend-making, etc. This makes it much easier to fiddle with your custom colors, which few of us can resist.
```{r}
jColors <- c('chartreuse3', 'cornflowerblue', 'darkgoldenrod1', 'peachpuff3',
'mediumorchid2', 'turquoise3', 'wheat4', 'slategray2')
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = jColors, main = 'custom colors!')
with(jDat, text(x = gdpPercap, y = lifeExp, labels = jColors,
pos = rep(c(1, 3, 1), c(5, 1, 2))))
```
### What colors are available? Ditto for symbols and line types
Who would have guessed that R knows about "peachpuff3"? To see the names of all `r length(colors())` the built-in colors, use `colors()`.
```{r}
head(colors())
tail(colors())
```
But it's much more exciting to see the colors displayed! Lots of people have tackled this -- for colors, plotting symbols, line types -- and put their work on the internet. Some examples:
* JB printed color names [on a white background](r.col.white.bkgd.pdf) and [on black](r.col.black.bkgd.pdf)
* JB printed [the first 30 plotting symbols](r.pch.pdf) (presumably using code found elsewhere or in documentation? can't remember whom to credit).
* In [Chapter 3 of R Graphics 1st edition](https://www.stat.auckland.ac.nz/~paul/RGraphics/chapter3.html), Paul Murrell shows predefined and custom line types in Figure 3.6 and plotting symbols in Figure 3.10.
* Earl F. Glynn offers [an excellent resource](http://research.stowers-institute.org/efg/R/Color/Chart/) on R's built-in named colors.
### RColorBrewer
Most of us are pretty lousy at choosing colors and it's easy to spend too much time fiddling with them. [Cynthia Brewer](http://en.wikipedia.org/wiki/Cynthia_Brewer), a geographer and color specialist, has created sets of colors for print and the web and they are available in the add-on package `RColorBrewer`. You will need to install and load this package to use.
```{r}
#install.packages("RColorBrewer")
library(RColorBrewer)
```
Let's look at all the associated palettes.
```{r fig.height = 9}
display.brewer.all()
```
They fall into three classes. From top to bottom, they are
* sequential: great for low-to-high things where one extreme is exciting and the other is boring, like (transformations of) p-values and correlations (caveat: here I'm assuming the only exciting correlations you're likely to see are positive, i.e. near 1)
* qualitative: great for non-ordered categorical things -- such as your typical factor, like country or continent. Note the special case "Paired" palette; example where that's useful: a non-experimental factor (e.g. type of wheat) and a binary experimental factor (e.g. untreated vs. treated).
* diverging: great for things that range from "extreme and negative" to "extreme and positive", going through "non extreme and boring" along the way, such as t statistics and z scores and signed correlations
You can view a single RColorBrewer palette by specifying its name:
```{r fig.height = 3}
display.brewer.pal(n = 8, name = 'Dark2')
```
The package is, frankly, rather clunky, as evidenced by the requirement to specify `n` above. Sorry folks, you'll just have to cope.
Here we revisit specifying custom colors as we did above, but using a palette from RColorBrewer instead of our artisanal "peachpuff3" work of art. As before, I display the colors themselves but you'll see we're not getting the friendly names you've seen before, which brings us to our next topic.
```{r}
jBrewColors <- brewer.pal(n = 8, name = "Dark2")
plot(lifeExp ~ gdpPercap, jDat, log = 'x', xlim = jXlim, ylim = jYlim,
col = jBrewColors, main = 'Dark2 qualitative palette from RColorBrewer')
with(jDat, text(x = gdpPercap, y = lifeExp, labels = jBrewColors,
pos = rep(c(1, 3, 1), c(5, 1, 2))))
```
### Hexadecimal RGB color specification
Instead of small positive integers and Crayola-style names, a more general and machine-readable approach to color specification is as hexadecimal triplets. Here is how the RColorBrewer Dark2 palette is actually stored:
```{r}
brewer.pal(n = 8, name = "Dark2")
```
The leading `#` is just there by convention. Parse the hexadecimal string like so: `#rrggbb`, where `rr`, `gg`, and `bb` refer to color intensity in the red, green, and blue channels, respectively. Each is specified as a two-digit base 16 number, which is the meaning of "hexadecimal" (or "hex" for short). Here's a table relating base 16 numbers to the beloved base 10 system.
```{r include = FALSE}
library(xtable)
foo <- t(cbind(hex = I(c(as.character(0:9), LETTERS[1:6])),
decimal = I(as.character(0:15))))
foo <- xtable(foo)
```
```{r results = 'asis', echo = FALSE}
print(foo, type='html')
```
Example: the first color in the palette is specified as "#1B9E77", so the intensity in the green channel is 9E. What does that mean?
$$
9E = 9 * 16^1 + 14 * 16^0 = 9 * 16 + 14 = 158
$$
Note that the lowest possible channel intensity is `00` = 0 and the highest is `FF` = 255.
Important special cases that help you stay oriented. Here are the saturated RGB colors, red, blue, and green:
```{r include = FALSE}
foo <- data.frame(colorName = c("blue", "green", "red"),
hex = c("#0000FF", "#00FF00", "#FF0000"),
red = c(0, 0, 255), green = c(0, 255, 0), blue = c(255, 0, 0))
foo <- xtable(foo, digits = 0)
```
```{r results = 'asis', echo = FALSE}
print(foo, type='html', include.rownames = FALSE)
```
Here are shades of gray:
```{r include = FALSE}
jIntensity <- c(255, 171, 84, 0)
foo <- data.frame(colorName = c("white, gray100", "gray67",
"gray33", "black, gray0"),
hex = c("#FFFFFF", "#ABABAB", "#545454", "#000000"),
red = jIntensity, green = jIntensity, blue = jIntensity)
foo <- xtable(foo, digits = 0)
```
```{r results = 'asis', echo = FALSE}
print(foo, type='html', include.rownames = FALSE)
```
Note that everywhere you see "gray" above, you will get the same results if you substitute "grey". We see that white corresponds to maximum intensity in all channels and black to the minimum.
To review, here are the ways to specify colors in R:
* a positive integer, used to index into the current color palette (queried or manipulated via `palette()`)
* a color name among those found in `colors()`
* a hexadecimal string; in addition to a hexadecimal triple, in some contexts this can be extended to a hexadecimal quadruple with the fourth channel referring to alpha transparency
Here are some functions to read up on if you want to learn more -- don't forget to mine the "See Also" section of the help to expand your horizons: `rgb()`, `col2rgb()`, `convertColor()`.
### Alternatives to the RGB color model, especially HCL
The RGB color space or model is by no means the only or best one. It's natural for describing colors for display on a computer screen but some really important color picking tasks are hard to execute in this model. For example, it's not obvious how to construct a qualitative palette where the colors are easy for humans to distinguish, but are also perceptually comparable to one other. Appreciate this: we can use RGB to describe colors to the computer __but we don't have to use it as the space where we construct color systems__.
Color models generally have three dimensions, as RGB does, due to the physiological reality that humans have three different receptors in the retina. ([Here is an informative blog post](http://manyworldstheory.com/2013/01/15/my-favorite-rgb-color/) on RGB and the human visual system.) The closer a color model's dimensions correspond to distinct qualities people can perceive, the more useful it is. This correspondence facilitates the deliberate construction of palettes and paths through color space with specific properties. RGB lacks this concordance with human perception. Just because you have photoreceptors that detect red, green, and blue light, it doesn't mean that your *perceptual experience* of color breaks down that way. Do experience the color yellow as a mix of red and green light? No, of course not, but that's the physiological reality. An RGB alternative you may have encountered is the Hue-Saturation-Value (HSV) model. Unfortunately, it is also quite problematic for color picking, due to its dimensions being confounded with each other.
What are the good perceptually-based color models? CIELUV and CIELAB are two well-known examples. We will focus on a variant of CIELUV, namely the Hue-Chroma-Luminance (HCL) model. It is written up nicely for an R audience in Zeileis, et al (see References for citation and link). There is a companion R package `colorspace`, which will help you to explore and exploit the HCL color model. Finally, this color model is fully embraced in `ggplot2` (as are the `RColorBrewer` palettes).
Here's what I can tell you about the HCL model's three dimensions:
* Hue is what you usually think of when you think "what color is that?" It's the easy one! It is given as an angle, going from 0 to 360, so imagine a rainbow donut.
* Chroma refers to colorfullness, i.e. how pure or vivid a color is. The more something seems mixed with gray, the lower its chromaticity. The lowest possible value is 0, which corresponds to actual gray. The maximum value varies with luminance.
* Luminance is related to brightness, lightness, intensity, and value. Low luminance means dark and indeed black has luminance 0. High luminance means light and white has luminance 1.
> Full disclosure: I have a hard time really grasping and distinguishing chroma and luminance. As we point out above, they are not entirely independent, which speaks to the weird shape of the 3 dimensional HCL space.
Figure 6.6 in Wickham's `ggplot2` book is helpful for understanding the HCL color space.
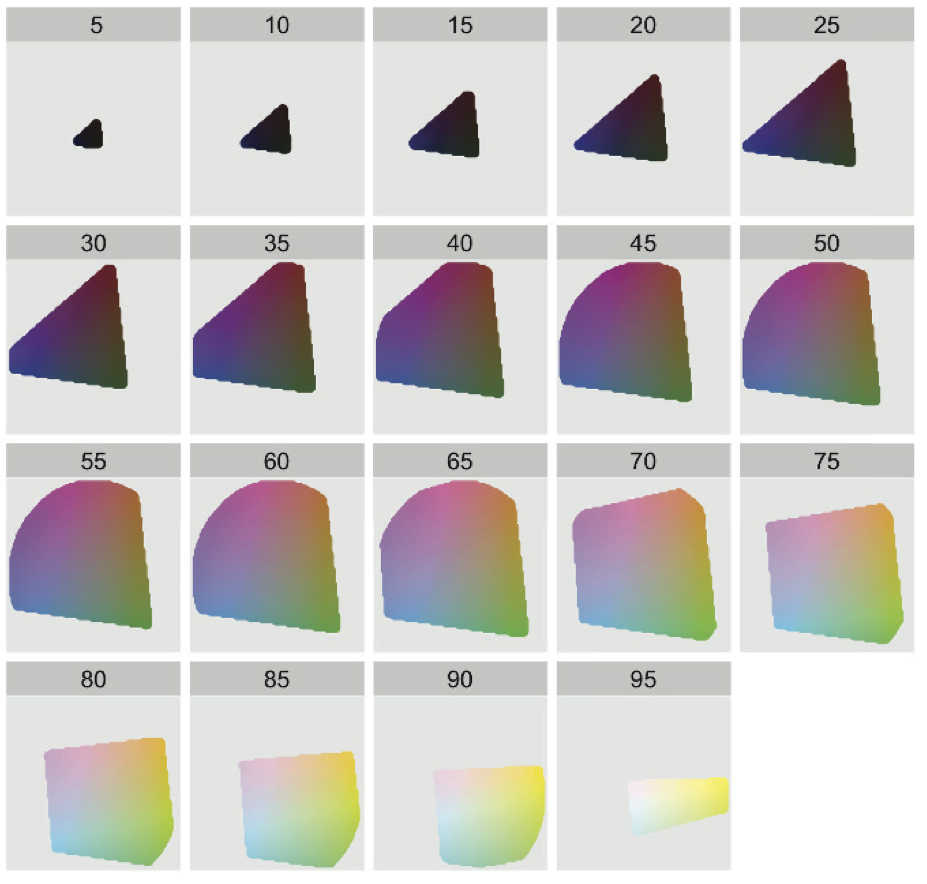
> JB re-phrasing and combining Wickham's description and caption for this figure: Each facet or panel depicts a slice through HCL space for a specific luminance, going from low to high. The extreme luminance values of 0 and 100 are omitted because they would, respectively, be a single black point and a single white point. Within a slice, the centre has chroma 0, which corresponds to a shade of grey. As you move toward the slice's edge, chroma increases and the color gets more pure and intense. Hue is mapped to angle.
A valuable contribution of the `colorspace` package is that it provides functions to create color palettes traversing color space in a rational way. In contrast, the palettes offered by `RColorBrewer`, though well-crafted, are unfortunately fixed.
> Here I plan to insert/recreate some visuals from the Zeileis et al paper or from the `colorspace` vignette. For the moment, that stuff is sitting in the PDF slides. So go there!
### Accomodating color blindness
the `dichromat` package ([on CRAN](http://cran.r-project.org/web/packages/dichromat/))
> obviously need to do some writing here!
### Clean up
```{r eval = FALSE}
## NOT RUN
## execute this if you followed my code for
## changing the default plot symbol in a simple, non-knitr setting
## reversing the effects of this: opar <- par(pch = 19)
par(opar)
```
### References
Achim Zeileis, Kurt Hornik, Paul Murrell (2009). Escaping RGBland: Selecting Colors for Statistical Graphics. Computational Statistics & Data Analysis, 53(9), 3259-3270. [DOI](http://dx.doi.org/10.1016/j.csda.2008.11.033) | [PDF](http://eeecon.uibk.ac.at/~zeileis/papers/Zeileis+Hornik+Murrell-2009.pdf)
[Vignette](http://cran.r-project.org/web/packages/colorspace/vignettes/hcl-colors.pdf) for the `colorspace` package
Earl F. Glynn (Stowers Institute for Medical Research)
* [excellent resources](http://research.stowers-institute.org/efg/R/Color/Chart/) for named colors, i.e. the ones available via `colors()`
* informative talk ["Using Color in R"](http://research.stowers-institute.org/efg/Report/UsingColorInR.pdf), though features some questionable *use* of color itself
Blog post [My favorite RGB color](http://manyworldstheory.com/2013/01/15/my-favorite-rgb-color/) on the Many World Theory blog
ggplot2: Elegant Graphics for Data Analysis [available via SpringerLink](http://ezproxy.library.ubc.ca/login?url=http://link.springer.com.ezproxy.library.ubc.ca/book/10.1007/978-0-387-98141-3/page/1) by Hadley Wickham, Springer (2009) | [online docs (nice!)](http://docs.ggplot2.org/current/) | [author's website for the book](http://ggplot2.org/book/), including all the code | [author's landing page for the package](http://ggplot2.org)
* Section 6.4.3 Colour
### Notes from the future
Consider incorporating in future version of this material:
* Example-laden article by Bernice E. Rogowitz and Lloyd A. Treinish of IBM Research ["Why Should Engineers and Scientists Be Worried About Color?"](http://www.research.ibm.com/people/l/lloydt/color/color.HTM), h/t [@EdwardTufte](https://twitter.com/EdwardTufte). Importance of signalling where zero is in colorspace, perceptually based color systems (they talk about hue, saturation, and luminance), when the heck was this written?
<div class="footer">
This work is licensed under the <a href="http://creativecommons.org/licenses/by-nc/3.0/">CC BY-NC 3.0 Creative Commons License</a>.
</div>