VarioML is a collaborative, community-driven specification in active development. Your first step should be to contact admin at varioml.org, as documentation is not yet complete.
Important:
- All new implementations should use validation to ensure consistency with the core specification. Contact admin at varioml.org for help with generating validation scripts using Schematron.
- Use standard ontologies for the content, such as the Sequence Ontology (http://www.sequenceontology.org/) and VariO (http://variationontology.org/).
- Some terms are defined using SKOS vocabularies: https://github.com/VarioML/VarioML/tree/master/ontology/skos.
- Source attributes in database xrefs (such as gene and ref_seq) as well as ontology terms should use database abbreviations defined in the MIRIAM registry (http://identifiers.org). For example, use hgnc.symbol for gene names, refseq for NCBI reference sequences, and obo.so for Sequence Ontology references.
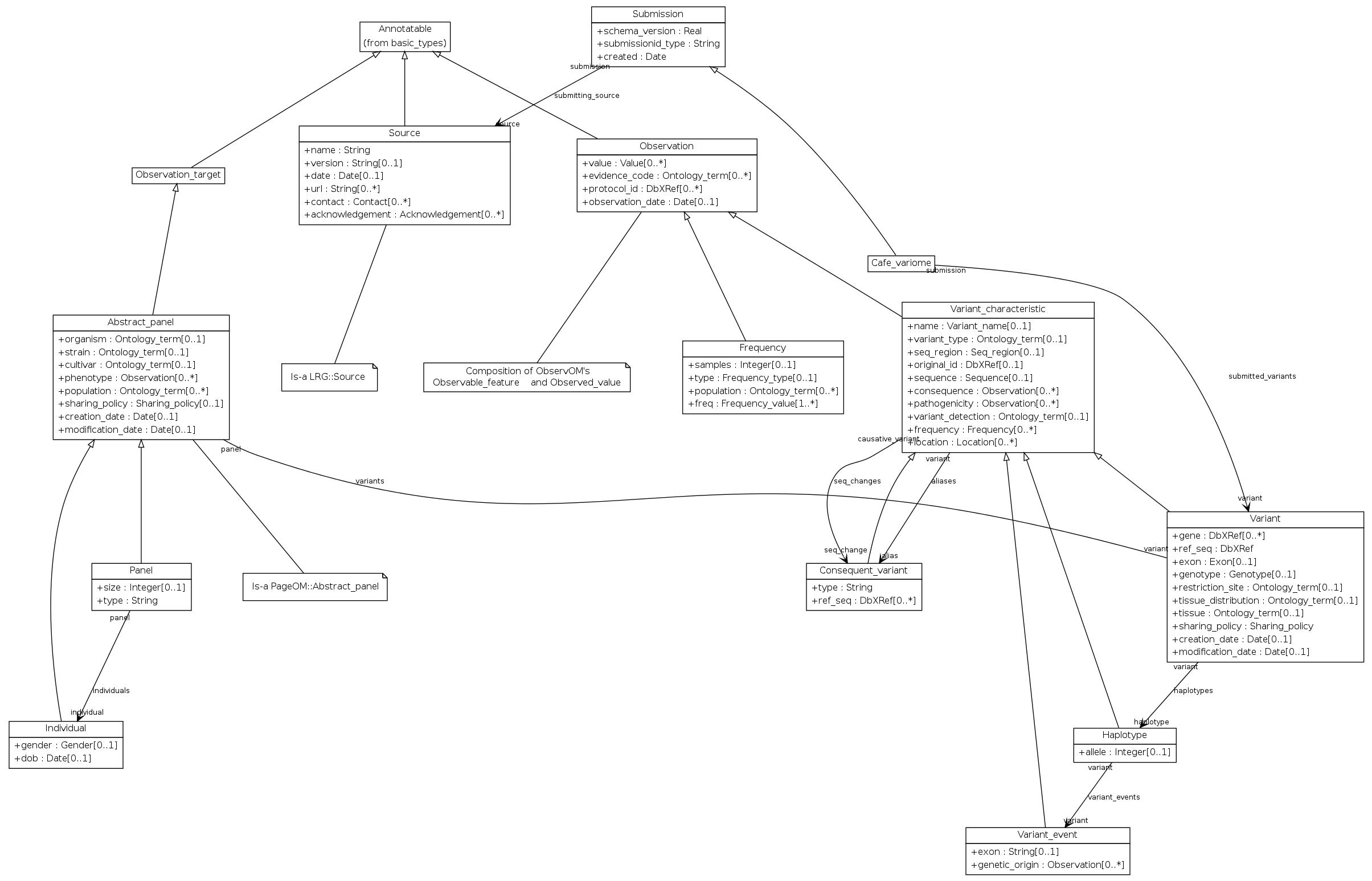
VarioML is currently implemented in Café Variome (http://cafevariome.org).
- The specification for this implementation is here: http://varioml.org/cafevariome_minspec.htm.
- A Schematron implementation is here: https://raw.github.com/VarioML/VarioML/master/xml/cafe_variome/cafe_variome_validator.sch
This project collects the source schema and validation tools for VarioML, a biomedical data standard supporting the integration, federation, and exchange of LSDB Data (http://varioml.org/). VarioML is desinged for curated LSDB data submissions and application integeration and is focused on individual variant entries. Format may not be suitable for handling bulk high throughput data, in which case e.g. VCF fomat may be more sutable.
These are shared here so biomedical researchers, clinicians, and bioinformaticians can:
- always have the current and canonical version of the data standard schemas and tools
- fork and modify a schema to adapt it to a specific research or clinical need
- log and track issues, and contributed improvements
Source schema, examples, and validation tools are grouped by implementation format of VarioML, with each format serving a large segment of biomedical users. E.g: the cafe_variome directory contains schema, examples, and validation tools for the Cafe Variome format, which facilitates the exchange of sequence variant data from diagnostic laboratories to diverse third parties (http://www.cafevariome.org/).
More information on VarioML can be found at:
- reference: http://varioml.org/
- wiki: http://www.gen2phen.org/groups/varioml
You can suggest improvements by entering an Issue, or if you see a technical improvement you can make yourself, by:
- cloning the repository
- making your changes
- checking your changes using validation tools such as those collected here: http://varioml.org/val.htm
- sending a pull request (http://help.github.com/pull-requests/)
This same process can be used to create a new schema for a specific research or clinical use.
VarioML has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement number 200754 - the GEN2PHEN project.
Demo applications can be found here https://github.com/VarioML/Apps
- 19.8.2012 Population changed to singleton in frequency. Do not have impact in existing XML implementations (property not used)
- 8.8.2012 Gene has changed to list of genes. One mutation may have impact on more than one genes
- 8.8.2012 EXI (http://www.w3.org/XML/EXI/) binary XML support added. EXI can reduce the size of disk and memory usage by 3-~10 times, already without compression.
- 9.2.2012: JAXB / JSON (based on Jacson) API implementation is in org.varioml.jaxb folder. Code is not fully tested. SimpleXML implementation will be retired (support is possible though)
- Note: Version 1.0 is now on a separate branch. The main trunk is heading towards release 2.0. The only difference between 1.0 and 2.0 is that the variant element can have haplotype elements containing variants which are in the cis position. Namespaces will be kept the same.
- Comments/feedback: admin <> varioml.org. Please send email if you are using the software so that we can accomodate your requirements!