ALTHOUGH I TAKE GREAT PAIN TO TRY TO REPRESENT WHAT IS BEING REPORTED TAKE THE OUTPUT OF THIS SIMULATOR WITH PLENTY OF SALT.
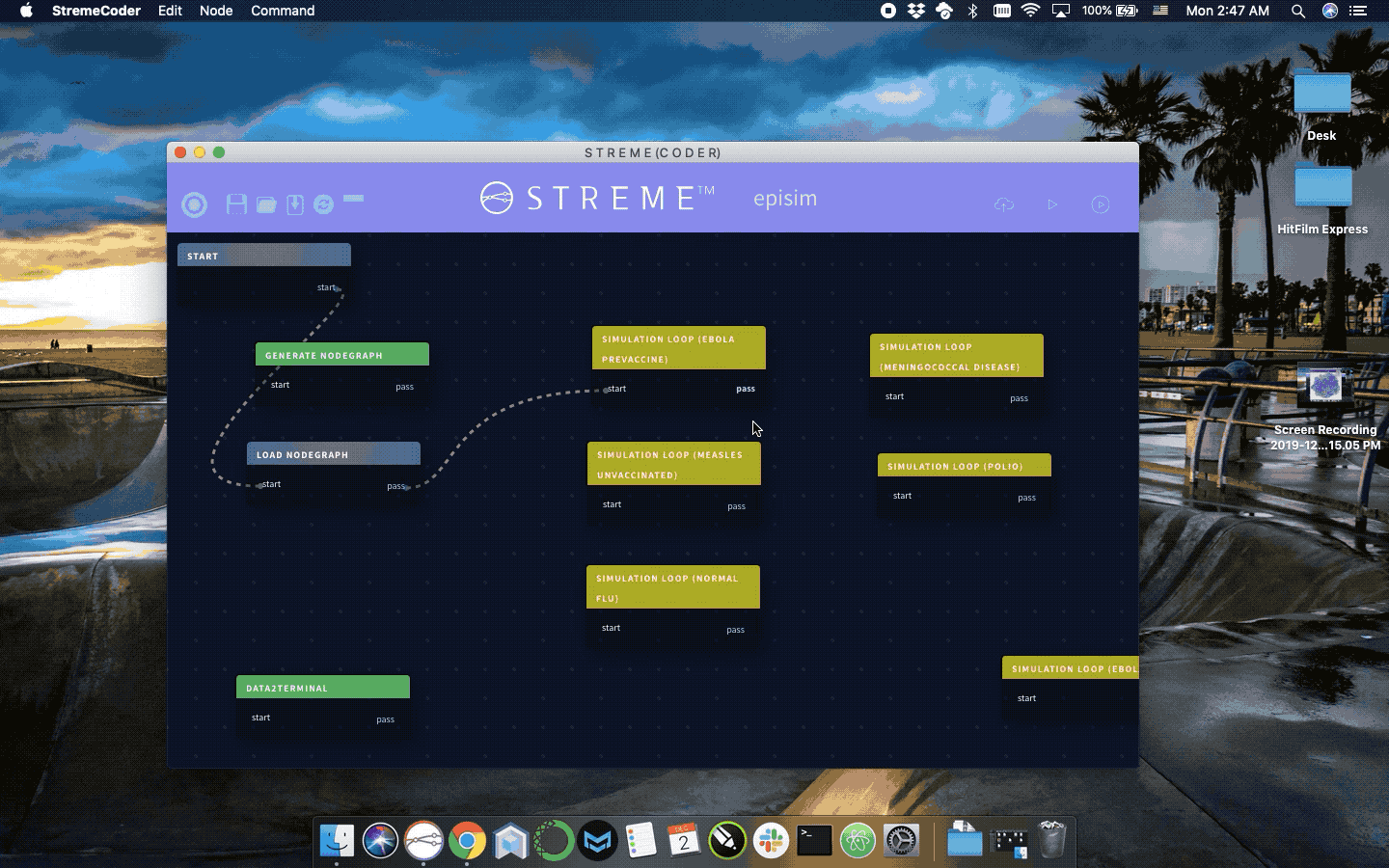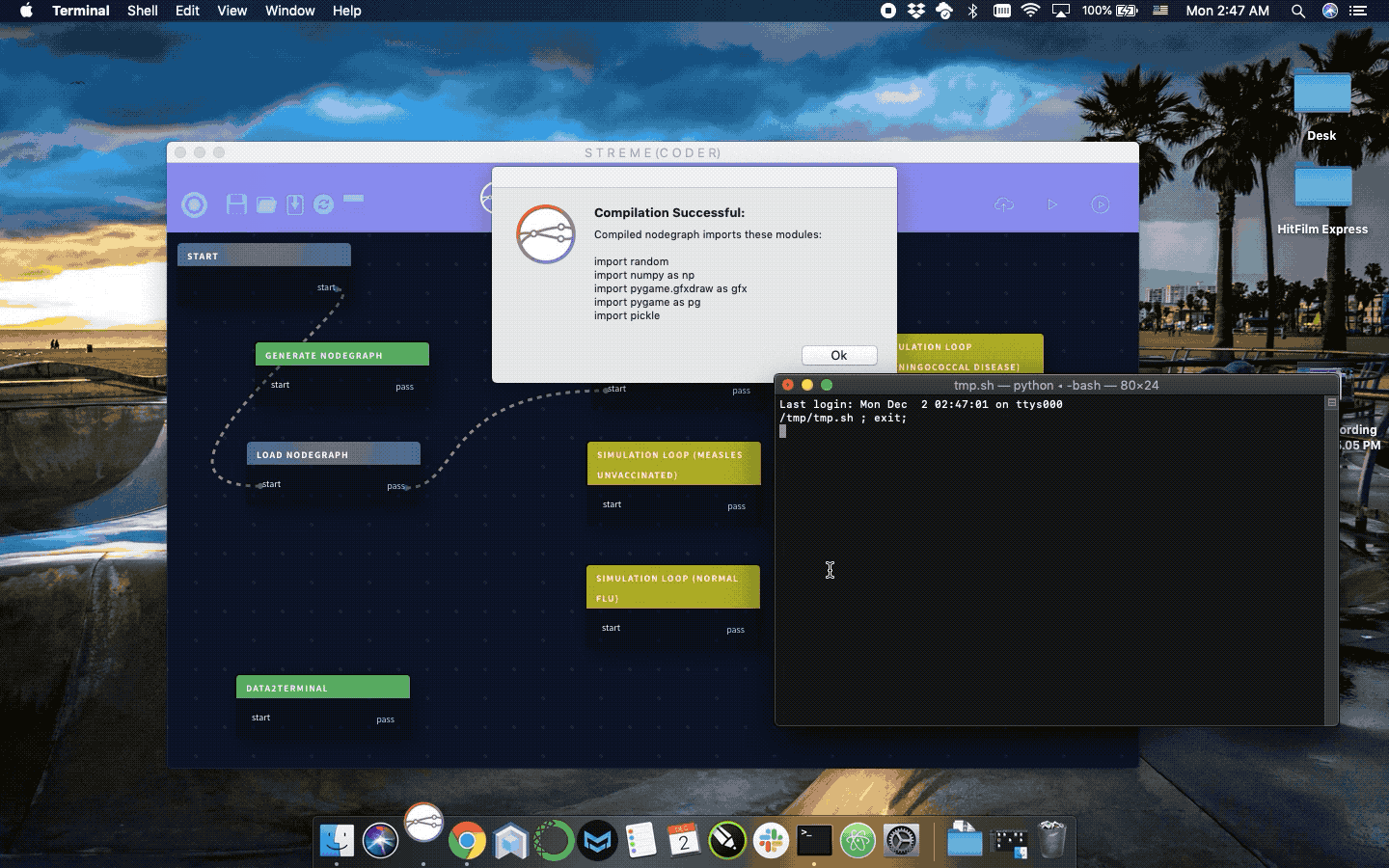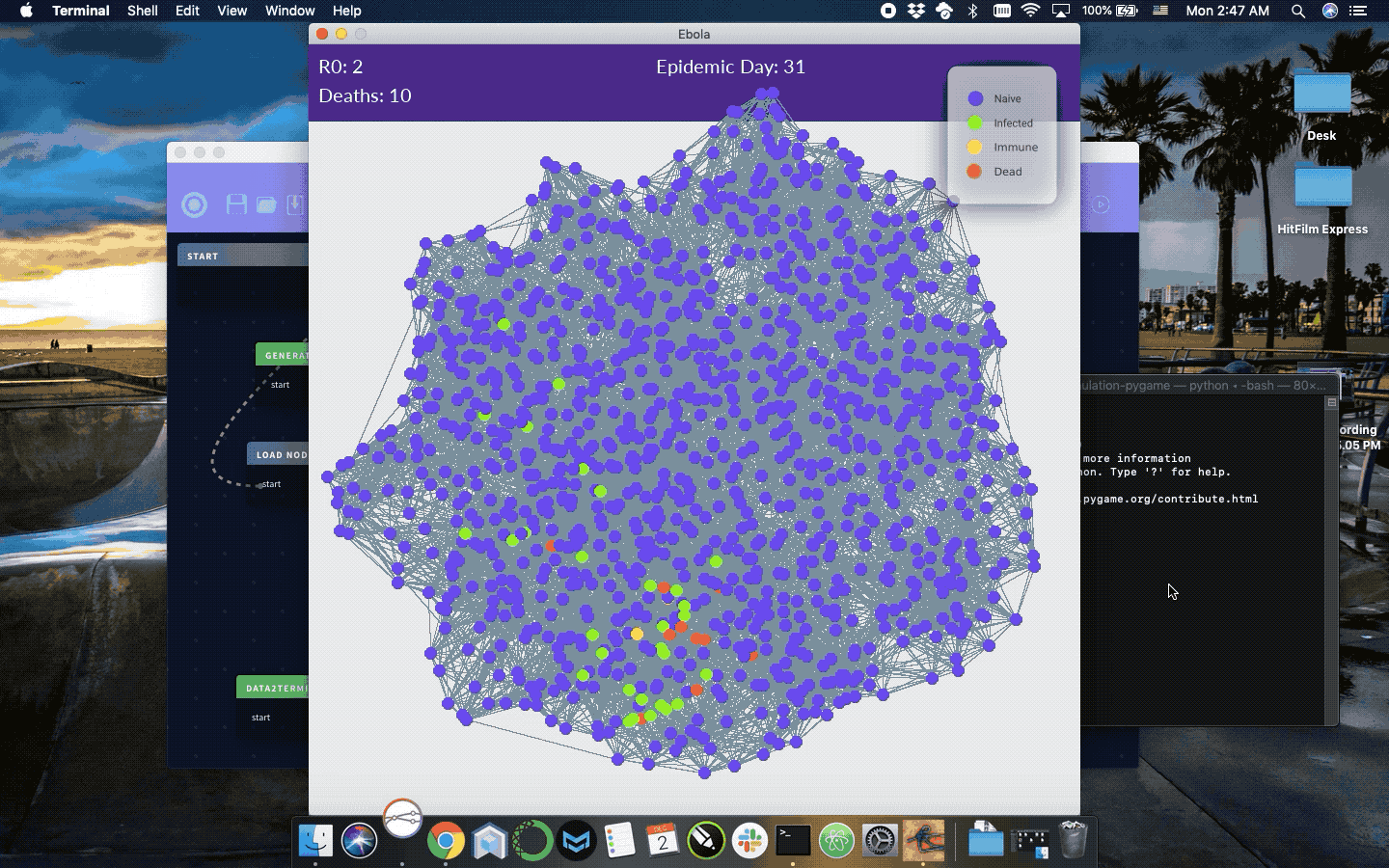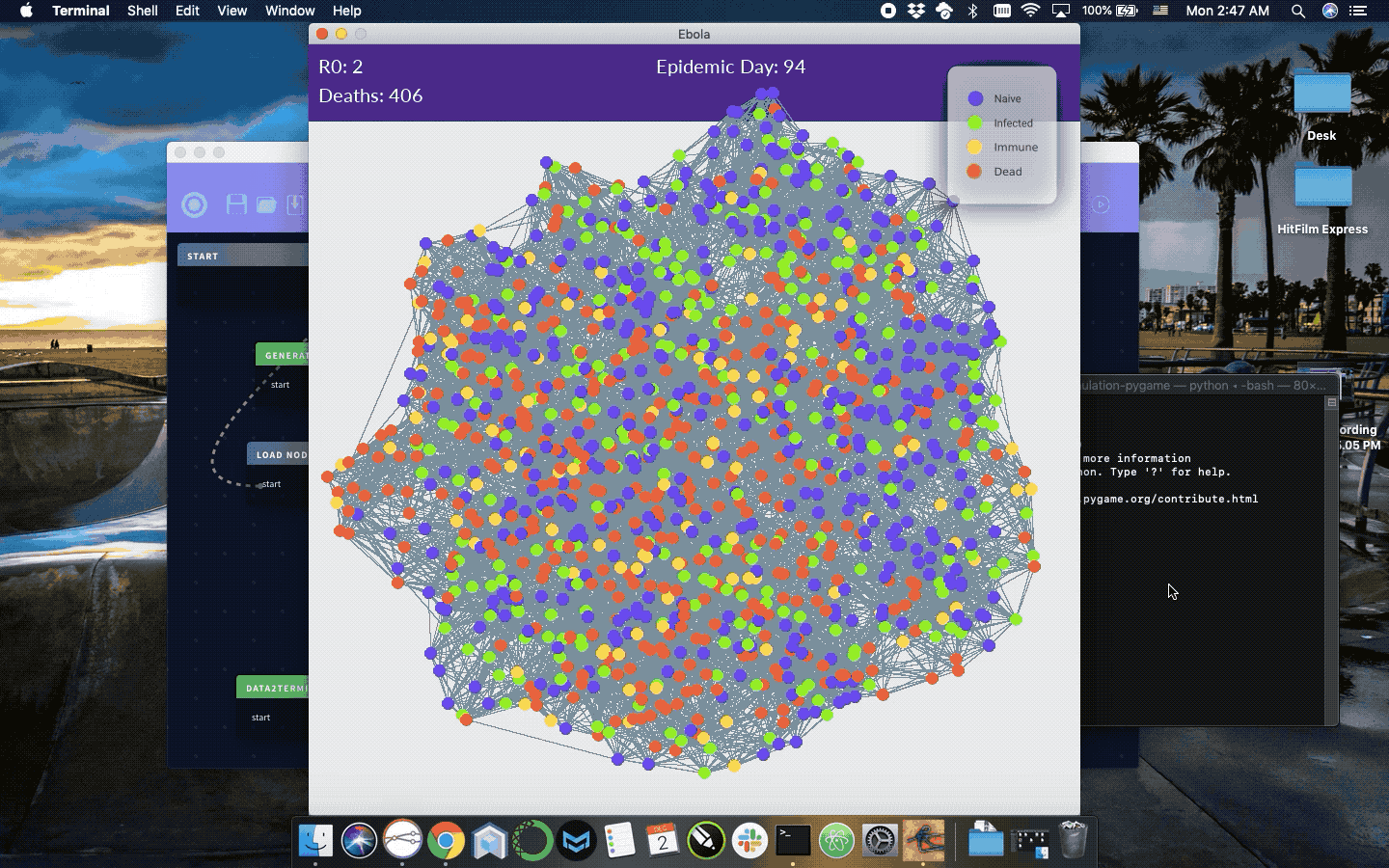
Simulation of infection progression given a reasonably natural network of connected individuals. StremeCoder Desktop is the IDE used to maintain this code and generate different infection parameters, but is not required and the python source can be modified directly in the simloop function to simulate different diseases.
You can also use the webversion for free StremeCoder Live it runs on your browser so it lacks some system functions that can only work on desktop apps, but it will still get the job done.
This simulation uses a social network generated in networkx
nx.newman_watts_strogatz_graph(nodes, k, prob)
where
nodes = 1200
k = 20
prob = 0.1
To reduce runtime and reproduce the same population for each disease node a pickle was generated of that random starter population and run through a pygame simulation node.
The simulation takes in
R0, infection probability, mortality rate, infection length, or infection probability if R0 is set to 0. Otherwise, infection probability is estimated from R0 algebraically. Each cycle of the simulation for each infected node connection infection and mortality are simulated by random number selection against provided probabilities. The simulation is set to end at 365 days. To use this simulation in a montecarlo remove the pygame dependencies to eliminate the visualizations and perform your desired statistical calculation on the resulting network then repeat the process n times.
This simulation requires python3, pygame, networkx, and numpy
Runs the visualization of the node state changes
pip install pygame
or
pip3 install pygame
To map the node network into the screen resolution space. Refer to SCIPY
conda install numpy
Generates the social network of nodes to run in the simulation You need the latest version or you will get an error
pip install networkx
or
pip3 install networkx
pip install seaborn
pip install pandas
In a terminal run
python episim.py