-
Notifications
You must be signed in to change notification settings - Fork 2
Simulation Framework
On the mark of every new year, the age of every alive organism in the ecosystem is incremented by 1.
New organisms are spawned at specific locations in the ecosystem based on the values in the current.json file of the species. For the very first generation, the locations are randomly selected otherwise the offspring are spawned right between their respective parents’ coordinates.
In case the birth is taking place because of one or more parents, the chromosome of the offspring is generated by exhaustively taking random bits from each parent to construct the final chromosome. In this way, the traits of parents are always passed down the generations.
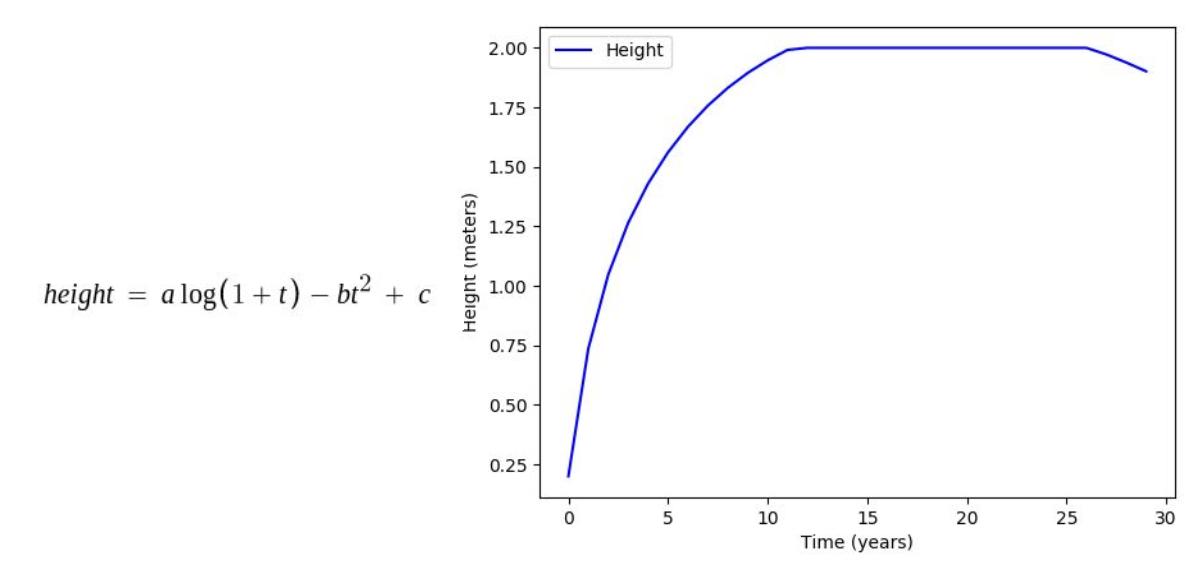
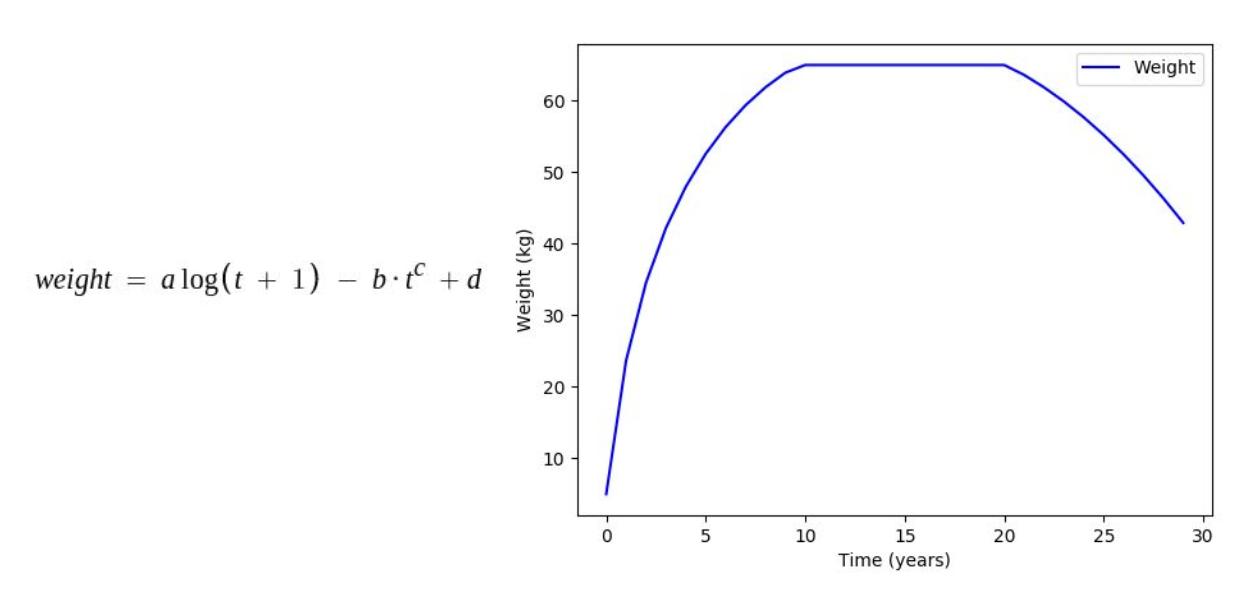
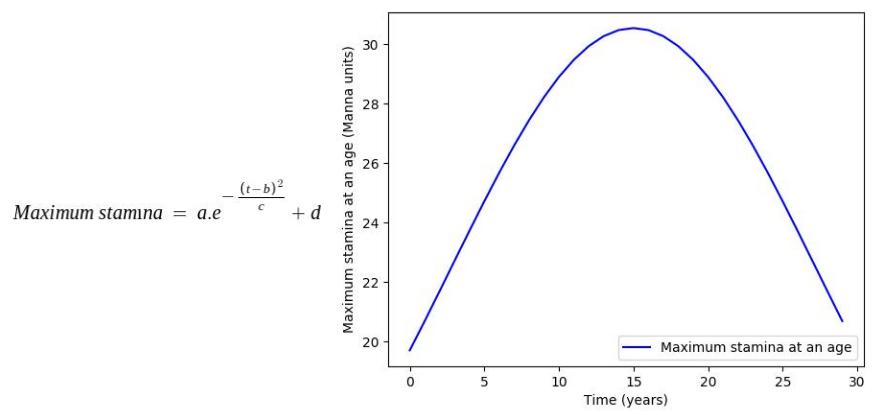
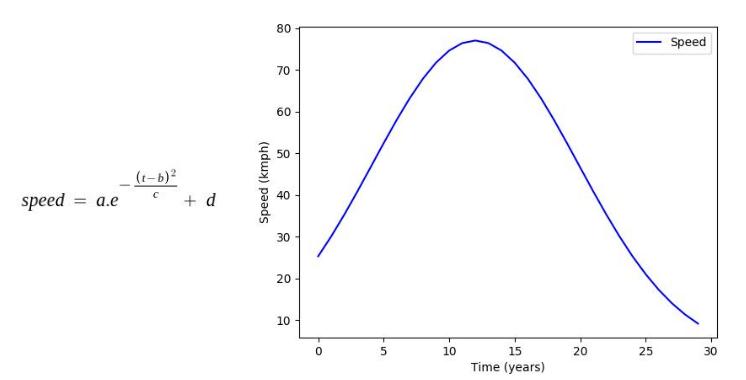
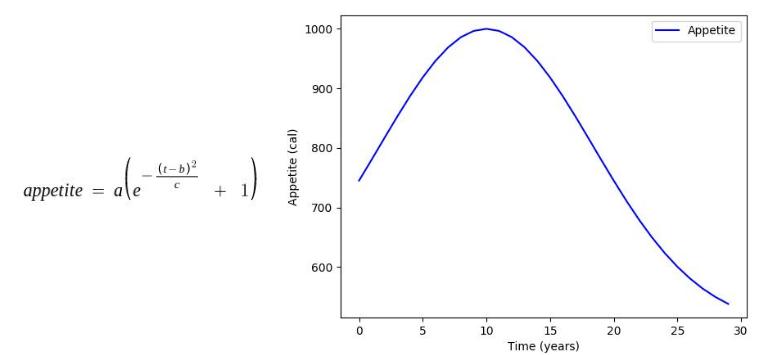
Factors such as height and weight are hugely dependent on the age by a predefined function mimicking the actual relationship between them in the real world. In this stage, all such parameters in each organism undergo the changes caused by their age increment in the previous stage.
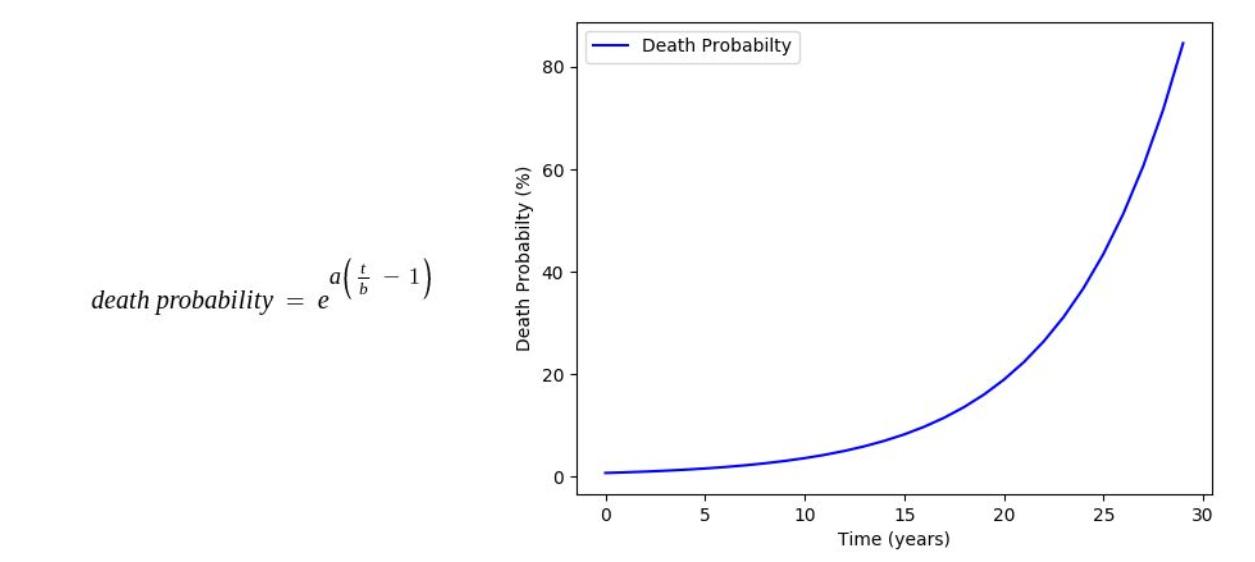
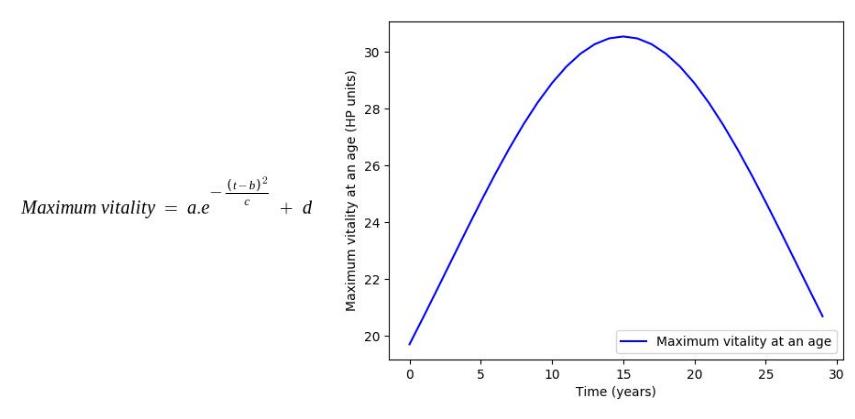
Some of the age dependencies are shown as follows:
Where, a, b, c, d, e are mathematical constants
For implementing Genetic Learning over the generations, a fitness factor is maintained throughout the lifespan of an organism determining its fitness to survive the next year in the simulation. Death of an organism is governed by two major factors:
a) Age
b) Fitness
The weightage of each of the above factors is used to determine the final death factor for an organism, which can be specified by the creator (user) in the base.json file. The relationship between death factor and fitness/age is a complex and probabilistic one with its nature tending towards a higher value of death factor for a higher age and lower fitness.
The organisms selected for death in the previous step are removed from the current organism buffer and also from the master database containing the list of currently alive organisms. No records for dead organisms are kept.
The mating activity can be divided into three significant phases:
a) Matchmaking
b) Conceiving
c) Mutation
Pairs of organisms are selected from each individual species. No inter-species cross-breeding is currently supported in the simulation. The selection of parents is done on the basis of their fitness value, such that individuals with higher fitness have a higher chance of being selected for matchmaking and with other individuals of similar fitness. This ensures that unfit organisms do not pollute the gene pool of fit organisms ultimately leading to the downfall of the species, strengthening the concept of Genetic Learning as seen in real life. In the case of asexual reproduction, the pairs are selected just on the basis of fitness irrespective of gender.
Each pair attempts to conceive offspring at this stage. The successful conceiving of offspring depends on factors governed by the creator (user) as defined in the base.json file. Also, the number of offspring generated per activity of conceiving depends on similar factors set by the creator(user). For example, animals have a higher chance of conceiving offsprings but a lower number of offspring per conceiving; plants have a lower chance of successful offsprings but a higher number of offsprings for successful conceiving.
For diversity in the gene pool, some of the genes during the creation of offspring are modified. Due to a rather simple binary representation of chromosomes in the simulation framework, adding mutation is as simple as flipping random bits based on a predefined mutation probability.
The entire Simulation Framework has been divided into a hierarchy of classes to make our implementation modular and easier to maintain. We have used a modified form of polymorphism which will be explained a bit later to gain runtime performance.
-
Entity - This class is at the topmost level of the class hierarchy. Any class which is a part of the simulation framework has to be a subclass of Entity.
-
Organism - This class is a direct subclass of Entity. Organism class has all the attributes and behaviour which are common to all organisms. Each organism has to be a subclass of this class.
-
Animal - This class is a subclass of Organism and has the attributes and behaviour which are common to Animals only. For example, attributes like stamina, appetite, gender are present in Animals but not Plants.
-
Plant - This class is yet another subclass of Organism and has behaviours which are common to Plants only.
-
-
-
God - This class represents the user who has the power to modify the ecosystem. Since there can be only ONE user there can be ONE God ONLY.
We have opted for a combination of static and dynamic polymorphism for high performance. Unlike the polymorphic design implemented in strictly Object Oriented languages like JAVA where polymorphism can only be implemented at runtime, in our C++ implementation the calls from Organism -> Animal and Organism -> Plant have all been resolved statically. In the case of Entity -> Organism, we have opted for dynamic polymorphism with pure virtual functions as the algorithms in God class are implemented in terms of an Entity whose type is not known at compile-time. The static polymorphism has been implemented using the well known Curiously Recurring Template Pattern (CRTP) which provides nearly 6x performance gains over dynamic polymorphism when compiler optimization flags and inlining is enabled.
-
The births and deaths of organisms belonging to a species occur on the same time of the year.
-
The fitness of an organism (or unique Chromosome) in a species is defined by a finite and exhaustive set of values or parameters (called Genes).
-
There will be a new generation of organisms every year without fail unless there are no suitable organisms to breed.
-
Organisms of the same species may crossbreed with their parent or offspring to contribute to the diversity in the gene pool.
-
An organism may survive multiple generations, however, the probability of death increases exponentially each passing unit of time.
🏠 About | 🔌 Setup | 🔩 Framework | 💻 Code